You are here:
stamms-lab.net >>
RNA-Bioinformatics
|
|
|
 |
 |
| A:
minigenes containing one cassette exon |
|
# |
name |
species |
minigene |
reference |
|
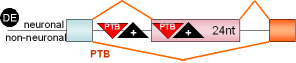
1 |
GABAAγ2
24nt Exon |
rat |
 |
 |
|
2 |
clathrin light chain B exon EN |
rat |
 |
 |
|
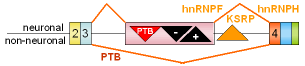
3 |
src
exon N1 |
mouse |
 |
 |
|
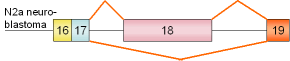
4 |
NCAM
exon 18 |
mouse |
 |
 |
|
5 |
MHC-B
exon N30 |
human |
 |
 |
|
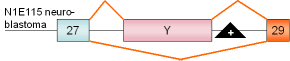
6 |
agrin
exon Y |
mouse |
 |
 |
|
7 |
tau,
exon 6 |
human |
 |
 |
|
8 |
fibronectin
EIIA |
mouse |
 |
 |
|
9 |
fibronectin
EIIB |
rat |
 |
 |
|
10 |
insulin
receptor
exon 11 |
human |
 |
 |
|
11 |
NCAM,
exon MSDb |
mouse |
 |
 |
|
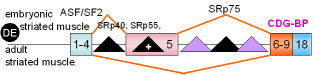
12 |
cTNT,
exon 5 |
chicken |
 |
 |
|
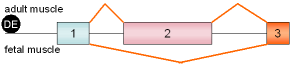
13 |
AMP
deaminase 1
exon 2 |
rat |
 |
 |
|
14 |
CD44,
exon 5 |
mouse |
 |
 |
|
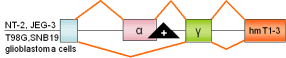
15 |
FGFR-1,
exon α |
human |
 |
 |
|
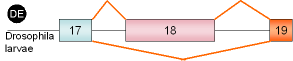
16 |
myosin
heavy chain,
exon 18 |
Droso-phila |
 |
 |
|
17 |
FcγRIIA
exon Tm |
human |
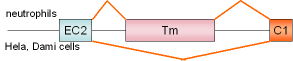 |
 |
|
18 |
interleukin-3α, exon 8 |
mouse |
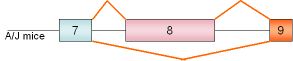 |
 |
|
19 |
DHFR, exon 2A, reporter gene |
hamster |
 |
 |
|
20 |
HIV-1,
exon 6D |
HIV-1 |
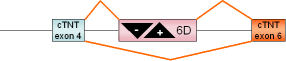 |
 |
|
21 |
SRp20, exon 4 |
human |
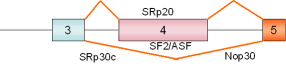 |
 |
|
22 |
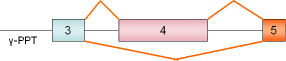
PPT, exon 4 |
rat |
 |
 |
B:
minigenes containing multiple cassette exons |
|
# |
name |
species |
minigene |
reference |
|
23 |
fast sceletal TnT, exons 4-7 |
rat |
 |
 |
|
24 |
CD45, exons 4-6 |
human, mouse |
 |
 |
|
25 |
APP, exons 7,8 |
human |
 |
 |
C:
minigenes containing mutually exclusive exons |
|
# |
name |
species |
minigene |
reference |
|
26 |
α-tropo-myosin exons 2,3 |
rat |
 |
 |
|
27 |
α-tropo-myosin exons
NM,SK |
human |
 |
 |
|
28 |
β-tropo-myosin exons
6A,B |
chicken |
 |
 |
|
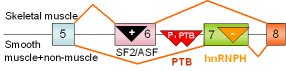
29 |
β-tropo-myosin exons 6,7 |
rat |
 |
 |
|
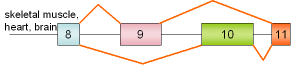
30 |
pyruvate-kinase M exons 9,10 |
human |
 |
 |
|
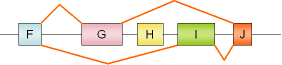
31 |
albumin, exons G,H |
rat |
 |
 |
|
32 |
MLC, exons 1/3 |
rat |
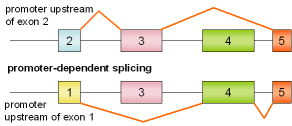 |
 |
|
33 |
FGFR,
K-SAM |
human |
 |
 |
D:
minigenes containing a retained intron |
|
# |
name |
species |
minigene |
reference |
|
34 |
bGH, intron D |
bovine |
 |
 |
E:
minigenes containing incremental combinatorial
exons |
|
# |
name |
species |
minigene |
reference |
|
35 |
tau, exons 2,3 |
human |
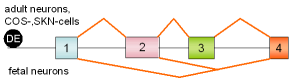 |
 |
F:
minigenes containing alternative 3' splice sites |
|
# |
name |
species |
minigene |
reference |
|
36 |
CT/CGRP |
human |
 |
 |
|
37 |
dsx RO |
Droso-phila |
 |
 |
|
38 |
M-tra |
Droso-phila |
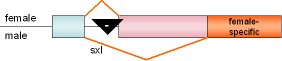 |
 |
|
39 |
BPV-1 |
BPV-1 |
 |
 |
G:
minigenes containing alternative 5' splice sites |
|
# |
name |
species |
minigene |
reference |
| 40 |
E1A |
Adeno-virus |
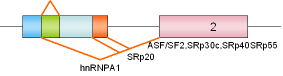 |
 |
| 41 |
SERCA2, exon 2a |
hamster |
 |
 |
| 42 |
Caldesomon gene |
human |
 |
 |
| 43 |
SWAP |
human |
 |
 |
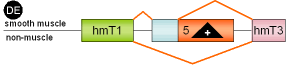
| 44 |
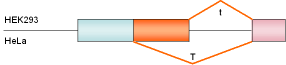
SC40
t-antigen |
SV40 |
 |
 |
| 45 |
β-globin, β-thalassemic allele |
human |
 |
 |
H:
minigenes containing alternative 3'- and 5'
splice sites |
|
# |
name |
species |
minigene |
reference |
|
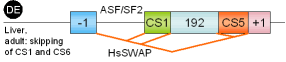
46 |
fibronectin, exon IIICS |
human |
 |
 |
|
 |
 |
|
| |
|
| |
|
|
|