|
Mechanism of alternative splicing
Splice site selection is regulated by the binding of trans-acting factors to RNA sequence elements. These factors work in a concentration dependent manner, which acts as a ?cellular code? that needs to be deciphered. The challenge in determining the regulation of splice sites is the high degeneracy of the regulatory sequences and the low specificity of the individual interactions. We therefore tackle this problem from two ends: very detailed work in model systems and genome-wide analysis. Both approaches generate theories that can be tested by the other method. To analyze alternative splicing on a genome-wide level, we established databases of alternative exons, their regulatory features and functions. Experimentally, we characterized several new splicing regulatory proteins and established model systems we can test both in vivo and in vitro. Several of these model systems (SMN2, tau and tra2-beta1) are relevant for human diseases, i.e. spinal muscular atrophy, Alzheimer?s disease and cancer. The advantage of the two-way approach is illustrated by this example: We identified a putative exonic enhancer in abnormal weak neuronal exons by bioinformatic means. Later, we could demonstrate that this enhancer is present in exon 10 of neurofilament tau, where it binds to the protein tra2-beta1 that we studied. A detailed analysis revealed that mutation of this enhancer causes frontotemporal dementia and that tra2-beta1 expression is changed in Alzheimer?s disease.
Splicing is catalysed by a multienzyme complex, the spliceosome.
We use the  yeast two-hybrid
system (A) to isolate novel splicing components through their
interaction with already known splicing factors. Using libraries made
from rat total brain, rat hippocampus and human HeLa cells, we found
novel yeast two-hybrid
system (A) to isolate novel splicing components through their
interaction with already known splicing factors. Using libraries made
from rat total brain, rat hippocampus and human HeLa cells, we found
novel  networks of spliceosomal
protein interactions (B). networks of spliceosomal
protein interactions (B). |
|
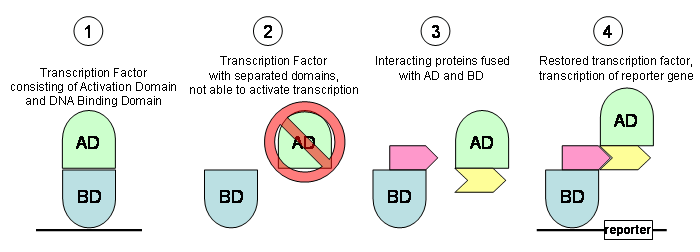
A transcription factor works by binding to a promotor sequence
using its DNA binding domain and activates polymerase II
transcription with its activation domain (1). In the yeast two hybrid
system, these two domains have been separated. Due to the separation
they cannot activate transcription (2). In the yeast two hybrid
system, the two domains are expressed as fusion proteins (3). For
example, two different spliceosomal proteins can be fused to the
activation and binding domain. If the spliceosomal proteins interact,
a functional transcription factor is restored and turns on a reporter
gene that is used for selection (4).
 |