|
You
are here:
stamms-lab.net >> Overview
>>
Signals regulating
alternative splicing in the brain
|
How is neuron-specific splicing involved
in creating molecular plasticity of the
brain? |
Signal transduction pathways regulating alternative splicing
The use of alternative exons often changes during development, or in response to outside stimuli. However, the pathways that transduce the signal to the splicing machinery remain to be established. In order to develop therapeutic approaches for diseases caused by wrong splice site selection, an understanding of these signal transduction pathways is necessary. We established several systems where we can stimulate cells or intact animals and test changes in splicing regulatory proteins. We found that phosphorylation of splicing factors are at the end-point of signal transduction pathways in all systems. The phosphorylation of regulatory factors changes their binding properties, resulting either in different RNP complexes forming on the pre-mRNA or in sequestration of splicing factors. As a result, splice site selection is changed. Currently, we continue to analyze the autoregulation of the tra2-beta system experimentally. We found that the phosphorylation of tra2-beta1 is regulated by protein phosphatase 1 and could demonstrate that through this regulation alternative splicing events are influenced through cAMP and cGMP levels that regulate protein phosphatase 1 activity. These findings let to novel drug candidates against spinal muscular atrophy that are currently evaluated in mouse models.
It is not clear how neurons change their molecular
composition after they were activated, e.g. by comunication with
other neurons or by receiving information from outside. We have
demonstrated that massive neuronal activity evoked by an epileptic
episode changes the alternative splicing patterns of
 a
splicing factor and alters its isoform distribution (from htra2-beta1
to htra2-beta3) (A), as well as the a
splicing factor and alters its isoform distribution (from htra2-beta1
to htra2-beta3) (A), as well as the
 splicing
patterns of various cellular genes (B),
e.g. clathrin light chain B. In vivo, we could demonstrate a splicing
patterns of various cellular genes (B),
e.g. clathrin light chain B. In vivo, we could demonstrate a
 regulation of
clathrin light chain B splicing by the tra2-beta isoforms
(C) and were able to regulation of
clathrin light chain B splicing by the tra2-beta isoforms
(C) and were able to
 investigate
signal transducution pathways leading to alternative splicing events (E). investigate
signal transducution pathways leading to alternative splicing events (E).
These experiments indicate that alternative
splicing patterns can change in response to neuronal activity. The
subsequent change of protein isoform composition might be an
important way to conway neuronal plasticity. We are currently
investigating
 how
signals can reach the spliceosome
(D). how
signals can reach the spliceosome
(D). |
|
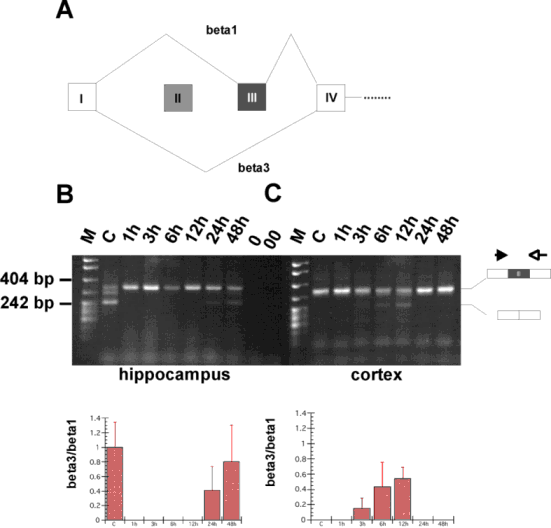
(A) Change of tra2-beta isoforms by neuronal
activity.
|
 |
|
RT-PCR using 1 µg total RNA from normal (C)
and stimulated (1h-48 h) hippocampus (left) and cortex (right). The
structure of the PCR products is schematically indicated on the
right. The band running between the 404 and 242 nt bands is most
likely a heterodimer of those two bands. 0: PCR with RNA, 00: PCR without template. (right) The same experiment was performed for
cortex.
|
|
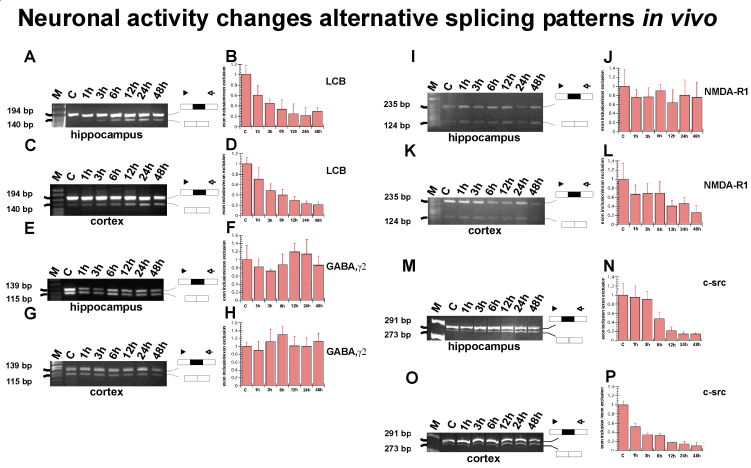
(B) Change of the splicing patterns of several
genes in response to neuronal activity.
|
 |
|
RT-PCR analysis of RNA from pilocarpine treated
hippocampus and cortex. Removal of tissue after pilocarpine injection
was at the time indicated. C: untreated control. A statistical
evaluation for three experiments is given on the right. Standard
deviations are indicated. Location of primers and cDNA structure are
schematically indicated for each gene. For each reaction, the ratio
of exon inclusion/exon exclusion was arbitrarily set to one in the
control tissue.
- (A) RT PCR for clathrin light chain B in hippocampus,
alternative exon EN
- (C) RT PCR for clathrin light chain B in cortex
- (E) RT PCR for GABAAg2-receptor, in hippocampus, alternative
exon 24
- (G) RT PCR for GABAAg2-receptor in cortex, alternative exon
24
- (I) RT PCR for NMDARI in hippocampus, alternative exon 21
- (K) RT PCR for NMDARI in cortex, alternative exon 21
- (M) RT PCR for c-src in hippocampus, alternative exon NI
- (O) RT PCR for c-src in cortex, alternative exon NI
|
|
(C)
Regulation of clathrin light chain B splicing by the tra2-beta isoforms
|
 |
|
|
|
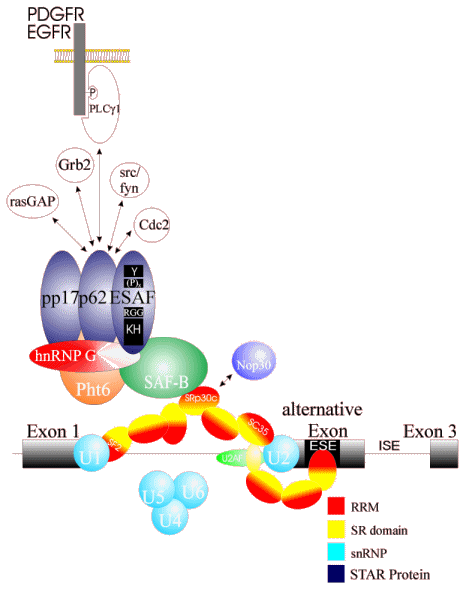
(D) Protein interactions leading from the
spliceosome to the outside world
|
 |
|
This picture shows the protein network identified by yeast-two
hybrid interactions and immunoprecipitations that could mediate a
signal form the surface of the cell to the nucleus.
|
(E)
Examples of signal transduction pathways that regulate
alternative splice site selection. |
|
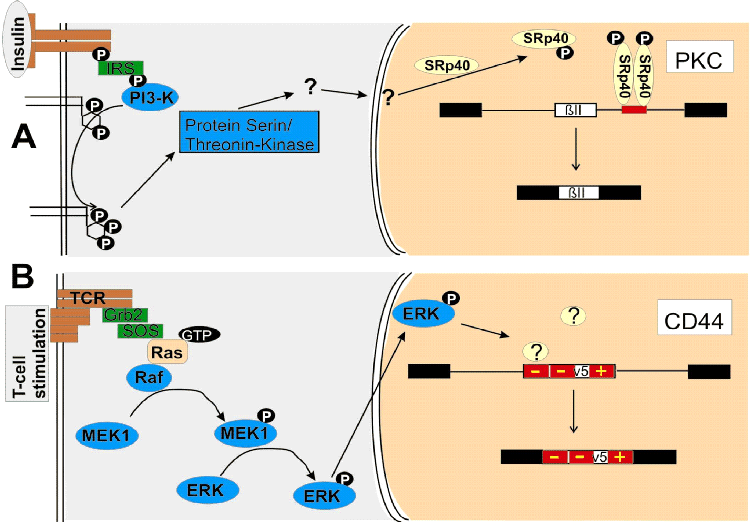
Examples of signal
transduction pathways that regulate alternative splice
site selection. The stimuli are shown in gray boxes on
the left. Receptors or ion channels are shown in brown.
The cytosol is shown in gray and the nucleus in sand
color. Kinases are shown in dark blue and splicing
regulatory proteins in yellow. Adapter molecules are
shown in green. Constitutive exons are shown as black
boxes and introns as lines. The alternative exons are
shown as white boxes. Regulatory RNA elements are shown
in red. The name of the gene is shown in a white box.
For simplicity, only one of the alternative mRNA
isoforms is shown. (A)
Regulation of PKC splicing by insulin. Binding of
insulin activates PI3-K via binding to the insulin
receptor substrate (IRS). PI3-K forms
phosphatidylinositol 3,4,5-trisphosphate, which
activates an unknown protein serine–threonine kinase
that ultimately phosphorylates SRp40, which acts on an
intronic element that leads to exon ßII inclusion. (B)
Regulation of CD44 after T-cell stimulation. Stimulation
of the T-cell receptor activates Ras via Grb2 (growth
hormone-binding protein 2) and Sos (‘son of sevenless’)
binding. Ras activates Raf, which phosphorylates MEK1,
which phosphorylates ERK. Phosphorylated ERK
translocates into the nucleus and releases repressors on
two silencing elements of the alternative exon v5. This
exon is subsequently included.
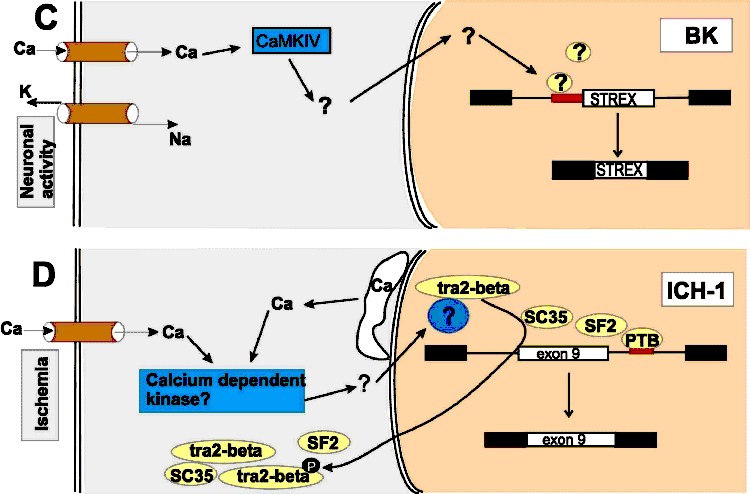 (C)
Regulation of the BK STREX exon by neuronal activity.
Neuronal activity opens voltage-gated calcium channels.
The rise in intracellular calcium activates CaMKIV,
which phosphorylates unknown proteins that bind to an
intronic element at the 3' splice site of the STREX exon.
De-repression of this element results in an inclusion of
the STREX exon. (D)
Regulation of the ICH-1
gene by ischemia. Ischemia evokes a rise in the
intracellular calcium levels by opening plasma membrane
calcium channels and releasing calcium stored in the
endoplasmatic reticulum (white structure). The rise in
calcium concentration probably activates a
calcium-dependent kinase, which ultimately leads to a
translocation of SR proteins from the nucleus to the
cytosol. The alternative exon of the
ICH-1
gene is repressed by the SR proteins SC35 and SF2,
possibly by indirect interaction with a pyrimidine-rich
repressing element downstream of the alternative exon.
The decrease in nuclear SR protein concentration results
in inclusion of the exon. (C)
Regulation of the BK STREX exon by neuronal activity.
Neuronal activity opens voltage-gated calcium channels.
The rise in intracellular calcium activates CaMKIV,
which phosphorylates unknown proteins that bind to an
intronic element at the 3' splice site of the STREX exon.
De-repression of this element results in an inclusion of
the STREX exon. (D)
Regulation of the ICH-1
gene by ischemia. Ischemia evokes a rise in the
intracellular calcium levels by opening plasma membrane
calcium channels and releasing calcium stored in the
endoplasmatic reticulum (white structure). The rise in
calcium concentration probably activates a
calcium-dependent kinase, which ultimately leads to a
translocation of SR proteins from the nucleus to the
cytosol. The alternative exon of the
ICH-1
gene is repressed by the SR proteins SC35 and SF2,
possibly by indirect interaction with a pyrimidine-rich
repressing element downstream of the alternative exon.
The decrease in nuclear SR protein concentration results
in inclusion of the exon.
|
|
|